OK OK. Everyone, everywhere keeps telling me "Viruses - don't forget about the viruses" whenever I talk about microbial communities or discuss work we are doing in my lab. Yes, I say "I know, I know" but then I panic and think "We can't even deal with the diversity of bacteria and archaea out there. We generally ignore the microbial eukaryotes. How the hell are we going to include viruses too?" But, in the end, if we are going to study microBIOMEs (as in, ecosystems that include viruses), then, well, we need to include all the players.
(I note - as I am writing this, my son asked me what I am doing and I said I am writing about viruses. He got excited and sad "Write about Ebola").
What got me on my viral kick today? Well, I have been thinking about viruses a lot recently. And then I saw this paper: The ISME Journal - Diversity of viral photosystem-I psaA genes. And it reminded me of the largely unknown diversity of viruses out there in the world.
Anyway - I was going to write a longer post about viruses, but I have to go and do something with my son.
Saturday, December 27, 2014
Friday, December 26, 2014
Paper of possible interest: Exploring interactions of plant microbiomes
Just got alerted to this new (open access) paper that seems like it should be of interest to those working on plant microbiomes Scientia Agricola - Exploring interactions of plant microbiomes. It has some useful summaries of work that has been done on plant microbiomes.
Wednesday, December 24, 2014
Do preprints count for anything? Not according to Elife & G3 & some authors ..
Well, just got pointed to this paper: Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms | eLife by Martial Marbouty, Axel Cournac, Jean-François Flot, Hervé Marie-Nelly, Julien Mozziconacci, Romain Koszul. Seems potentially really interesting.
It is similar in concept and in many aspects to a paper we published in PeerJ earlier in the year (see Beitel et al., 2014 Beitel CW, Froenicke L, Lang JM, Korf IF, Michelmore RW, Eisen JA, Darling AE. (2014) Strain- and plasmid-level deconvolution of a synthetic metagenome by sequencing proximity ligation products. PeerJ 2:e415 http://dx.doi.org/10.7717/peerj.415.
Yet despite the similarities to our paper and to another paper that was formally published around the time of ours, this new paper does not mention these other pieces of work any where in the introduction as having any type of "prior work" relevance. Instead, they wait until late in their discussion:
Taking advantage of chromatin conformation capture data to address genomic questions is a dynamic field: while this paper was under review, two studies were released that also aimed at exploiting the physical contacts between DNA molecules to deconvolve genomes from controlled mixes of microorganisms (Beitel et al., 2014; Burton et al., 2014).Clearly, what they are trying to do here is to claim that since they paper was submitted before these other two (including ours) was published, that they should get some sort of "priority" for their work. Let's look at that in more detail. Their paper was received May 9, 2014. Our paper was published online May 27 and the other related paper by Burton et al. was published online May 22. In general, if a paper on what your paper is about comes out just after you submit your paper, while your paper is still in review, the common, normal thing to be asked to do is to rewrite your paper to deal with the fact that you were, in essence, scooped. But that does not really appear to be the case here. They are treating this in a way as "oh look, some new papers came out at the last minute and we have commented on them." The last minute would be in this case, 6 months before this new paper was accepted. Seems like a long time to treat this as "ooh - a new paper came up that we will add a few comments about".
But - one could quibble about the ethics and policies of dealing with papers that were published after one submitted one's own paper. From my experience, I have always had to do major rewrites to deal with such papers. But maybe E-Life has different policies. Who knows. But that is where things get really annoying here. This is because it was May 27 when our FINAL paper came out online at PeerJ. However, the preprint of the paper was published on February 27, more than two months before their paper was even submitted. So does this mean that the authors of this new paper do not believe that preprints exist? It is pretty clear on the web site for our paper that there is a preprint that was published earlier. Given what they were working on - something directly related to what our preprint/paper was about, one would assume they would have seen it with a few simple Google searches. Or a reviewer might have pointed them to it. Maybe not. I do not know. But either way, our preprint was published long before their paper was submitted and therefore I believe they should have discussed it in more detail.
Is this a sign that some people believe preprints are nothing more than rumors? I hope not. Preprints are a great way to share research prior to the delays that can happen in peer review. And in my opinion, preprints should count as prior research and be cited as such. I note - the Burton group in their paper in G3 also did not reference our preprint in what I consider to be a reasonable manner. They add some comments in their acknowledgements
While this manuscript was in preparation, a preprint describing a related method appeared in PeerJ PrePrints (Beitel et al. 2014a). Note added in proof: this preprint was subsequently published (Beitel et al. 2014b).Given that our preprint was published before their paper was submitted too, I believe that they also should have made more reference to it in their paper. But again, I can only guess that both the Burton and the Marbouty group just do not see preprints as being respectable scientific objects. That is a bad precedent to set and I think the wrong one too. And it is a shame. A preprint is a publication. No - it is not peer reviewed. But that does not mean it should not be considered part of the scientific literature in some way. I note - this new paper from the Marbouty group seems really interesting. Not sure I want to dig into it any deeper if they are going to play games with the timing of submission vs. published "papers" as part of how they are positioning themselves to be viewed as doing something novel.
Tuesday, December 23, 2014
Buzzfeed includes microbiomes in "77 facts" but alas, gets the facts (and the math) wrong
Well, I admit it - I clicked on this Buzzfeed link someone posted on Facebook - 77 Facts That Sound Like Huge Lies But Are Actually Completely True. There are some pretty funny and interesting things on the list. However, I note - I did not click on the link per se for entertainment. I clicked on it to see if there was anything about microbes on the list. And happily, there was. Alas, what there was, was, well, a bit wrong:
Let's start with #67. "There is 10 times more bacteria in your body than actual body cells". Well, alas, this is a nice bit of information. But it is not based on facts. See Peter Andrey Smiths wonderful article in the Boston Globe about this issue. The Buzzfeed article links to a 2010 Discover Magazine bit about this topic. So they were trying to use facts. But alas, that was not based on facts itself.
Then, let's go to #68. "And 90% of the cells that make us up of aren’t human but mostly fungi and bacteria.". So - I am at a bit of a loss on this. First, isn't this really just rehashing #67 with the addition of fungi to the story? Also - I note the math here is weird. #67 would lead one to conclude that 90.9% of the cells in your body are bacterial (10:1 = 10/11 = 90.9%). So then if one adds fungi to the picture the percent of cells in our bodies that are not human goes DOWN to 90%. How does that work exactly?
So not only do they get the 10x as many cells fact wrong, they do something really weird by basically repeating #67 and then doing some weird math.
Oh well, glad they have something on microbiomes. Too bad it came out a bit wrong.
Let's start with #67. "There is 10 times more bacteria in your body than actual body cells". Well, alas, this is a nice bit of information. But it is not based on facts. See Peter Andrey Smiths wonderful article in the Boston Globe about this issue. The Buzzfeed article links to a 2010 Discover Magazine bit about this topic. So they were trying to use facts. But alas, that was not based on facts itself.
Then, let's go to #68. "And 90% of the cells that make us up of aren’t human but mostly fungi and bacteria.". So - I am at a bit of a loss on this. First, isn't this really just rehashing #67 with the addition of fungi to the story? Also - I note the math here is weird. #67 would lead one to conclude that 90.9% of the cells in your body are bacterial (10:1 = 10/11 = 90.9%). So then if one adds fungi to the picture the percent of cells in our bodies that are not human goes DOWN to 90%. How does that work exactly?
So not only do they get the 10x as many cells fact wrong, they do something really weird by basically repeating #67 and then doing some weird math.
Oh well, glad they have something on microbiomes. Too bad it came out a bit wrong.
Sunday, December 21, 2014
Crosspost - papers for sale
Crossposting from ICIS Blog (even though this is short - always good to reveal where things are posted first).
Well, this is much more elaborate than I could ever have imagined: For Sale: “Your Name Here” in a Prestigious Science Journal – Scientific American. Seems that there are services out there to help people write, in essence, bogus scientific papers filled with pithy somewhat reasonable sounding phrases about certain topics. Seems we could all use some more comprehensive full text analyses of papers to try and flag such activities.
Well, this is much more elaborate than I could ever have imagined: For Sale: “Your Name Here” in a Prestigious Science Journal – Scientific American. Seems that there are services out there to help people write, in essence, bogus scientific papers filled with pithy somewhat reasonable sounding phrases about certain topics. Seems we could all use some more comprehensive full text analyses of papers to try and flag such activities.
Thursday, December 18, 2014
Great idea from Nicole King - Lists of Women Speakers - more examples wanted
Just been pointed to this compilation from Nicole King (a brilliant professor at Berkeley): Lists of Women Speakers | The King Lab.
She has put together a collection of a few links:
She has put together a collection of a few links:
As a resource for those who are interested in increasing the number of female speakers at their scientific conferences, seminar series, etc., I offer here a compendium of female speakers from diverse fields. I've started with lists that I could find easily, but if you know of additional lists, please feel free to send along the links. I would also be happy to maintain similar lists for underrepresented minorities if any such lists exist.These are the lists she maintained:
General
- University of Minnesota, Distinguished Scientists and Engineers from Underrepresented Groups
- Women scientists that give awesome seminars
- Celebrating Women in Science Symposium
- Indiana University Joan Wood Lecture Series
Field-specific lists
- American Society of Cell Biology WICB Speaker Referral Service
- Neuroscience: Annie's List
- Theoretical/computational chemistry, material science, and biochemistry
- Synthetic Biology
- Prominent ecologists
- Geoscientists
- American Physical Society Women Speakers List
Are people aware of any other relevant lists like this? Please share I will compile ...
Wednesday, December 10, 2014
New lab paper: The microbes we eat: abundance and taxonomy of microbes consumed in a day’s worth of meals for three diet types
A new paper out from my lab (with Jenna Lang as the 1st author and in collaboration with Angela Zivcovic from the UC Davis Food For Health Initiative and the Department of Nutrition): The microbes we eat: abundance and taxonomy of microbes consumed in a day’s worth of meals for three diet types. The work in the paper focuses on characterizing the abundance and taxonomy of microbes in food from three model diets.
Basically, Angela prepared meals for these three diets
Lots of interesting things reported in the paper (read it, I insist). I note - this is a demonstration project in a way - trying to get the community and others to think about the source pools of microbes that come into our system from our food. It is by no means comprehensive or conclusive (read the caveats section of the paper). Congrats to Jenna and Angela for all their hard work. Anyway - the paper is Open Access in PeerJ. Eat it up.
UPDATE: Some press and blog coverage
Basically, Angela prepared meals for these three diets
Food was purchased and prepared in a standard American home kitchen by the same individual using typical kitchen cleaning practices including hand washing with non-antibacterial soap between food preparation steps, washing of dishes and cooking instruments with non-antibacterial dish washing detergent, and kitchen clean-up with a combination of anti-bacterial and non-antibacterial cleaning products. Anti-bacterial products had specific anti-bacterial molecules added to them whereas “non-antibacterial” products were simple surfactant-based formulations. The goal was to simulate a typical home kitchen rather than to artificially introduce sterile practices that would be atypical of how the average American prepares their meals at home. All meals were prepared according to specific recipes (from raw ingredient preparation such as washing and chopping, to cooking and mixing).And then she blended them and we characterzied the microbial communities in the blended samples:
After food preparation, meals were plated on a clean plate, weighed on a digital scale (model 157W; Escali, Minneapolis, MN), and then transferred to a blender (model 5,200; Vita-Mix Corporation, Cleveland, OH) and processed until completely blended (approximately 1–3 min). Prepared, ready to eat foods that were purchased outside the home were simply weighed in their original packaging and then transferred to the blender. 4 mL aliquots of the blended meal composite were extracted from the blender, transported on dry ice and then stored at −80 °C until analysis. The following analyses were completed using these meal composite samples: (1) total aerobic bacterial plate counts, (2) total anaerobic bacterial plate counts, (3) yeast plate counts, (4) fungal plate counts, and (5) 16S rDNA analysis for microbial ecology.And Jenna Lang coordainted the sequence analysis and then Angela and Jenna (with some help here and there from me) coordianted the analysis of the different microbial data and the writing of the paper.
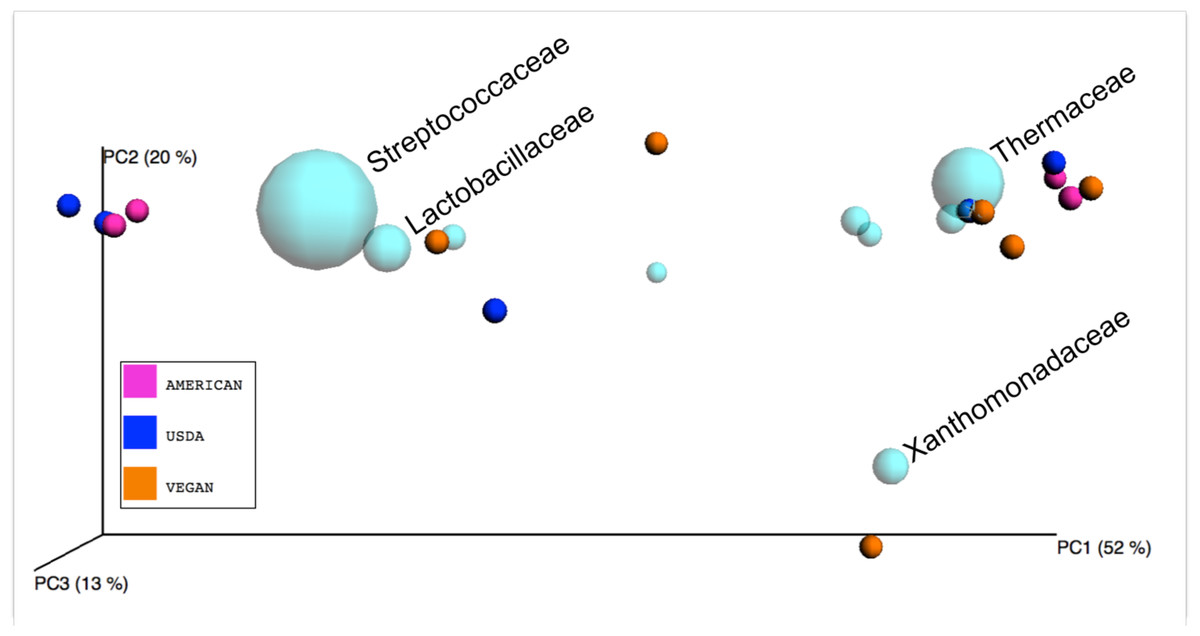 | ||
| Figure 5: Biplot of taxa in sample PCoA space. |
Lots of interesting things reported in the paper (read it, I insist). I note - this is a demonstration project in a way - trying to get the community and others to think about the source pools of microbes that come into our system from our food. It is by no means comprehensive or conclusive (read the caveats section of the paper). Congrats to Jenna and Angela for all their hard work. Anyway - the paper is Open Access in PeerJ. Eat it up.
UPDATE: Some press and blog coverage
- Kevin Bonham on SciAM Blogs: The Microbes in Your Kitchen (Or in your Starbucks mocha)
- Maddie Stone on Motherboard: Here’s How Many Microbes You Eat in a Single Day
- “This paper will hopefully convince the powers that be that the next study, actually looking at how different foods or diets with differing microbes affect people, should be funded soon,” Zivkovic told me.
- Nathan Gray: The microbes we eat: Study investigates bacteria in different diets
- Ross Pomeroy on Real Clear Science: This Is How Many Microbes You'll Eat Today
- Improbable Research: What’s Eating You, and/or Vice Versa: Microbes
- Marissa Fessenden at Smithsonian: You Eat Millions...Even Billions!...of Microbes Every Day
Saturday, December 06, 2014
Some history of hype regarding the human genome project and genomics
Just taking some notes here - relates to a discussion going on online. Would love pointers to other references relating to hype and the human genome project (including references that think it was not overhyped). I note - see some of my previous posts about this issue including: Human genome project oversold? sure but lets not undersell basic science and various Overselling Genomics awards.
Here are some things I have found:
White House press conference on announcing completetion of the human genome
The Human Genome Project: Hype meets reality
NOVA: Nature vs. Nurture Revisited
The human genome project, 10 years in: Did they oversell the revolution? in the Globa and Mail by Paul Taylor referring to: "Deflating the Genomic Bubble"
Also see Genomic Medicine: Too Great Expectations? by PP O Rourke
Also Has the Genomic Revolution Failed?
And Human genome 10th anniversary. Waiting for the revolution.
Science communication in transition: genomics hype, public engagement, education and commercialization pressures.
The Medical Revolution in Slate.
A Decade Later, Genetic Map Yields Few New Cures in the New York Times.
NNB report: Ten years later, Harvard assesses the genome map where regarding Eric Lander:
From Great 15-Year Project To Decipher Genes Stirs Opposition in the Times June 1990
From SCIENTIST AT WORK: Francis S. Collins; Unlocking the Secrets of the Genome
From READING THE BOOK OF LIFE: THE DOCTOR'S WORLD; Genomic Chief Has High Hopes, and Great Fears, for Genetic Testing June 2000 in the NY Times
The story goes through some predictions Francis Collins made for the future in a talk. These included:
Here are some things I have found:
White House press conference on announcing completetion of the human genome
Genome science will have a real impact on all our lives -- and even more, on the lives of our children. It will revolutionize the diagnosis, prevention and treatment of most, if not all, human diseases. In coming years, doctors increasingly will be able to cure diseases like Alzheimer's, Parkinson's, diabetes and cancer by attacking their genetic roots. In fact, it is now conceivable that our children's children will know the term cancer only as a constellation of stars.Collins et al. New Goals for the U.S. Human Genome Project: 1998–2003
The Human Genome Project (HGP) is fulfilling its promise as the single most important project in biology and the biomedical sciences— one that will permanently change biology and medicine.Human Genome -The Biggest Sellout in Human History
The Human Genome Project: Hype meets reality
NOVA: Nature vs. Nurture Revisited
After a decade of hype surrounding the Human Genome Project, punctuated at regular intervals by gaudy headlines proclaiming the discovery of genes for killer diseases and complex traits, this unexpected result led some journalists to a stunning conclusion. The seesaw struggle between our genes (nature) and the environment (nurture) had swung sharply in favor of nurture.
The human genome project, 10 years in: Did they oversell the revolution? in the Globa and Mail by Paul Taylor referring to: "Deflating the Genomic Bubble"
Also see Genomic Medicine: Too Great Expectations? by PP O Rourke
Also Has the Genomic Revolution Failed?
And Human genome 10th anniversary. Waiting for the revolution.
Science communication in transition: genomics hype, public engagement, education and commercialization pressures.
The Medical Revolution in Slate.
A Decade Later, Genetic Map Yields Few New Cures in the New York Times.
In announcing on June 26, 2000, that the first draft of the human genome had been achieved, Mr. Clinton said it would “revolutionize the diagnosis, prevention and treatment of most, if not all, human diseases.”
At a news conference, Francis Collins, then the director of the genome agency at the National Institutes of Health, said that genetic diagnosis of diseases would be accomplished in 10 years and that treatments would start to roll out perhaps five years after that.
Bursting the genomics bubble - Nature 2010
Has decoding the genome lived up to hype? BBC - 2010
The ethics of expectations: biobanks and the promise of personalised medicine.
The ethics of expectations: biobanks and the promise of personalised medicine.
NNB report: Ten years later, Harvard assesses the genome map where regarding Eric Lander:
At the same time, , he said genomic research has “gone so much faster than I would have imagined.” He cited " an explosion of work that will culminate, I think in the next five years, in a pretty comprehensive list of all the target that lead to different kinds of cancers and give us a kind of roadmap for finding the Achilles heel of cancers for therapeutics and diagnostics."while at the same time he blamed the press for the hype
From Great 15-Year Project To Decipher Genes Stirs Opposition in the Times June 1990
'Our project is something that we can do now, and it's something that we should do now,'' said Dr. James D. Watson, a Nobel laureate who heads the National Center for Human Genome Research at the National Institutes of Health. ''It's essentially immoral not to get it done as fast as possible.''
- Note the article has many complaining about the hype in the genome project even then ..
From SCIENTIST AT WORK: Francis S. Collins; Unlocking the Secrets of the Genome
And, Dr. Collins adds, there is nothing more important in science and medicine than the project he heads
Dr. Collins predicts that within 10 years everyone will have the opportunity to find out his or her own genetic risks, to know if cancer or heart attacks or diabetes or Alzheimer's disease, for example, lies in the future.
From READING THE BOOK OF LIFE: THE DOCTOR'S WORLD; Genomic Chief Has High Hopes, and Great Fears, for Genetic Testing June 2000 in the NY Times
The story goes through some predictions Francis Collins made for the future in a talk. These included:
- BY 2010, the genome will help identify people at highest risk of particular diseases, so monitoring efforts can focus on them.
- In cancer, genetic tests will identify those at highest risk for lung cancer from smoking. Genetic tests for colon cancer will narrow colonoscopy screening to people who need it most. A genetic test for prostate cancer could lead to more precise use of the prostate specific antigen, or P.S.A., test by identifying those men in whom the cancer is most likely to progress fastest. Additional genetic tests would guide treatment of breast and ovarian cancer.
- Three or four genetic tests will help predict an individual's risk for developing coronary artery disease, thus helping to determine when to start drugs and other measures to reduce need for bypass operations.
- Tests predicting a high risk for diabetes should help encourage susceptible individuals to exercise and control their weight. Those at higher risk might start taking drugs before they develop symptoms.
- BY 2020, doctors will rely on individual genetic variations in prescribing new and old drugs and choosing the dose. Pharmaceutical companies will take a second look at some drugs that were never marketed, or were taken off the market, because some people who took them suffered adverse reactions. It will take many years to develop such drugs and tests.
- Cancer doctors will use drugs that precisely target a tumor's molecular fingerprint. One such gene-based designer drug, Herceptin, is already marketed for treating advanced breast cancer.
- The genome project holds promise for the mental health field. ''One of the greatest benefits of genomic medicine will be to unravel some biological contributions to major mental illnesses like schizophrenia and manic depressive disease'' and produce new therapies, Dr. Collins said.
Friday, December 05, 2014
Some notes on "Citations for Sale" about King Abdulaziz University offerring me $$ to become an adjunct faculty
There is a news story in the Daily Cal titled "Citations for Sale" by Megan Messerly about King Abdullah University King Abdulaziz University trying to pay researchers who are highly cited to become adjunct professors there to boost their rankings. This article stemmed from a blog post by Lior Pachter. I was interviewed by the Daily Cal reporter about this because I had sent Lior some of the communications I had had with people from KAU where they tried to get me to do this.
I am posting here some of the email discussions / threads that I shared with Lior and Megan.
Thread #1.
Here is one thread of emails in which KAU tried to get me to become an Adjunct Professor. I have pulled out the text of the emails and removed the senders ID just in case this would get him in trouble.
Received this email 3/6/14
Dear Prof Jonathan,
How are you? I hope every thing is going well. I hope to share work with you and initiate a collaboration between you and me in biology department, Kind Abdulaziz university . My research focuses on (redacted detail). I hope that you would agree .
Redacted Name,Ph.DMy response:
Assistant Professor, Faculty of Sciences, King Abdulaziz University, Jeddah, KSA.
What kind of collaboration are you imagining?His response:
Hi Prof Jonathan,
Let me to explain that the king abdulaziz university initiated a project which is called Highly Cited Professor ( HiCi). This project can provide a contract between you and our university and from this contract you can get 7000 US$ as a monthly salary .So this project will allow you to generate two research proposal between you and a research stuff here in order to make an excellent publications in high quality journals as you always do.
I hope that I was clear. I' m looking forward to hear from you. Finally, I think that a very good chance to learn from you.Another email from him:
Dear prof Jonathan,
I' d like to tell that Prof Inder Verma will come tomorrow to our university as a highly cited professor and he also signed a contract with us. At March 28 Prof Paul Hebert will come to our university and we actually generated two projects with Prof Paul. I hope you trust me and you call Prof Inder and Paul to be sure.From me:
I trust you - just am very busy so not sure if this is right for meFrom him:
Sent from my iPhone
You will come to our university for just two visits annually and each visit will take only one week. Take you time to think. ByeAnother email from him
Seat Dr Jonathan,
What is your decision?
Monday, November 24, 2014
A night with Matt Groening and the importance of faeces, feces and faces
So - I participated in a fundraiser for Emily Levine's "The Edge of Chaos" film a week ago. And one of the key guests was Matt Groening. Not only did I get to hang out with him and discuss fecal transplants with him (really) but I had a front row seat to Matt discussing the history of how he came up with the general outline of the Simpson's characters.
And in addition to this being just awesome to witness, one part of it struck me. See the video below and in particular the part that struck me was the beginning:
Groening
basically said that only a few simple changes in faces can be recognized by people very easily. This reminds me of Jenna Lang's talk at the Lake Arrowhead meeting this year where she discussed using facial drawings as a form of visualizing microbiome data.
So - since I discussed fecal transplants with Matt and since he gave a good description of facial characteristics being easy to identify, I think we should definitely (1) try and get him to include microbiomes on the Simpsons and (2) for our work we should use Simpsons characters as model faces for different microbiomes ...
Oh and I also showed Groening some of the pics of my kids reading his "Hell" books:
And in addition to this being just awesome to witness, one part of it struck me. See the video below and in particular the part that struck me was the beginning:
Groening
basically said that only a few simple changes in faces can be recognized by people very easily. This reminds me of Jenna Lang's talk at the Lake Arrowhead meeting this year where she discussed using facial drawings as a form of visualizing microbiome data.
Citizen Microbiology: Opportunities for Engagement and Data Visualization, Lake Arrowhead, 2014 from jennomics
So - since I discussed fecal transplants with Matt and since he gave a good description of facial characteristics being easy to identify, I think we should definitely (1) try and get him to include microbiomes on the Simpsons and (2) for our work we should use Simpsons characters as model faces for different microbiomes ...
Oh and I also showed Groening some of the pics of my kids reading his "Hell" books:
So - basically it was a night of feces, faeces and faces. Seems ideal.
Sunday, November 23, 2014
Whole issues of Genome Biology/Genome Medicine on "Genomics of Infectious Disease"
Wow this has really got some nice papers: BioMed Central | Article collections | Genomics of infectious diseases special issue. I note - this goes well as a follow up to the series I co-coordinated in PLOS a few years back: Genomics of Emerging Infectious Disease - PLOS Collections
From their site:
Infectious diseases are major contributors to global morbidity and mortality, and have a devastating impact on public health. The World Health Organization estimates that 1 in 3 deaths worldwide are due to an infectious disease, with a disproportionate number occurring in developing regions.
While the completion of the first genome sequence of a pathogen, Haemophilus influenzae, in 1995 took decades of work, in recent years, high-throughput technologies have revolutionized the study of pathogens. Whole-genome sequences are now achievable within days and available for multiple pathogens, including those that cause neglected tropical diseases, which has advanced our understanding of the biology and evolution of pathogens. Crucially, such research has enabled important advances in the clinical management of infectious diseases, and continues to guide public health interventions worldwide.
In this cross-journal special issue, guest edited by George Weinstock (The Jackson Laboratory for Genomic Medicine, USA) and Sharon Peacock (University of Cambridge, UK), Genome Biology and Genome Medicine take stock of where we are now, with a collection of primary research and commissioned articles that discuss different aspects of the genomics of infectious diseases in human populations, including the progress made towards their eradication, and the remaining challenges in terms of both fundamental science and clinical management.I have copied the list from their site (I am pretty sure this is OK since these are #OpenAccess journals but not 100% sure):
Saturday, November 22, 2014
Strangest microbial headline of month: Bacteria on Russian ‘sex satellite’ survive reentry
 There is really not much to say other than to point everyone to this article: Bacteria on Russian ‘sex satellite’ survive reentry | Science | The Guardian
There is really not much to say other than to point everyone to this article: Bacteria on Russian ‘sex satellite’ survive reentry | Science | The GuardianIt defininely wins the strangest microbial headline of the month. The article restates some of the silly claims about how what they are finding supports panspermia .. but ignore the article and just enjoy the headline.
Moore Foundation: Request for Expressions of Interest: Increasing the Potential of Marine Microeukaryotes as Experimental Model Systems through the Development of Genetic Tools
Got this from the Gordon and Betty Moore Foundation and they said I could post it here.
Request for Expressions of Interest: Increasing the Potential of Marine Microeukaryotes as Experimental Model Systems through the Development of Genetic Tools
Marine Microbiology Initiative Gordon and Betty Moore Foundation November 21, 2014
The Marine Microbiology Initiative (MMI) at the Gordon and Betty Moore Foundation aims to enable scientists to uncover the principles that govern the interactions among microbes and that influence nutrient flow in the marine environment. MMI is targeting closing gaps in and supporting the advancement of experimental model systems in microbial oceanography to enable new ways to uncover fundamental biological mechanisms.
We are soliciting expressions of interest (EOIs) for early-stage research projects to develop methods to genetically manipulate marine microeukaryotes as a first step in breaking current bottlenecks in the advancement of experimental model systems. MMI has two primary foci for this expression of interest:
MMI encourages EOIs from “inter-organismal” teams of researchers – i.e., complementary groups that have experience in a well-established model system and with a microeukaryote that is not currently genetically tractable – whose collaborative effort will bring innovative approaches to the field.
MMI invites you to send an expression of interest via email that briefly outlines a research project (one paragraph or less), using the following template:
The opportunities that best align with MMI’s strategies and goals will be invited to submit proposals. MMI has allocated $7–10M to support this effort and anticipates making multiple, 2–3 year awards beginning in mid- 2015.
Please submit your EOI by Tuesday January 6, 2015 to Samantha Forde at samantha.forde@moore.org.
Request for Expressions of Interest: Increasing the Potential of Marine Microeukaryotes as Experimental Model Systems through the Development of Genetic Tools
Marine Microbiology Initiative Gordon and Betty Moore Foundation November 21, 2014
The Marine Microbiology Initiative (MMI) at the Gordon and Betty Moore Foundation aims to enable scientists to uncover the principles that govern the interactions among microbes and that influence nutrient flow in the marine environment. MMI is targeting closing gaps in and supporting the advancement of experimental model systems in microbial oceanography to enable new ways to uncover fundamental biological mechanisms.
We are soliciting expressions of interest (EOIs) for early-stage research projects to develop methods to genetically manipulate marine microeukaryotes as a first step in breaking current bottlenecks in the advancement of experimental model systems. MMI has two primary foci for this expression of interest:
-
Development of genetic tools for diatoms. Diatoms are key players in the world’s oceans, generating
~20% of the world’s organic carbon, and a strong community of researchers is in place suggesting
broad use of successfully developed methods. We are specifically interested in projects to develop
reverse and/or forward genetics.
-
Screening laboratory-scale culture collections for transformable marine microeukaryotes.
MMI encourages EOIs from “inter-organismal” teams of researchers – i.e., complementary groups that have experience in a well-established model system and with a microeukaryote that is not currently genetically tractable – whose collaborative effort will bring innovative approaches to the field.
MMI invites you to send an expression of interest via email that briefly outlines a research project (one paragraph or less), using the following template:
- The lead researcher’s name, institution, and expertise.
- Indication of focus on genetic tools for diatoms (category 1 above) or laboratory culture screening for transformability (category 2 above).
- For category 1, the name of the organism(s); or, for category 2, the taxonomic group(s) to be screened.
- A methodological or technical challenge that is hindering the development of a genetically manipulable marine microeukaryotic system that is ripe for solving and how you would address this challenge (3-5 sentences).
- The research team that would tackle this challenge, and why each team member’s expertise is relevant (one sentence per team member; please include institutional affiliations).
The opportunities that best align with MMI’s strategies and goals will be invited to submit proposals. MMI has allocated $7–10M to support this effort and anticipates making multiple, 2–3 year awards beginning in mid- 2015.
Please submit your EOI by Tuesday January 6, 2015 to Samantha Forde at samantha.forde@moore.org.
Post-doc w/ me, Jessica Green, Jay Stachowicz, and Jenna Lang on seagrass microbiomes
Postdoctoral Position in Microbial Ecology and Evolution
Jessica Green at the University of Oregon Green (http://pages.uoregon.edu/green/) is currently seeking a postdoctoral researcher to explore fundamental questions in microbial ecology and evolution. Applicants should have a PhD in a biological, computational, mathematical, or statistical field with extensive training using theory and/or modeling to understand the ecology and evolution of complex biological communities, and strong writing skills. Experience developing and applying quantitative phylogenetic ecological methods is highly desirable, but not explicitly required for candidates who have otherwise demonstrated strong quantitative skills.
The successful candidate will play a key role in the Seagrass Microbiome Project (http://seagrassmicrobiome.org) in collaboration among Jonathan Eisen https://phylogenomics.wordpress.com), Jay Stachowicz http://www-eve.ucdavis.edu/stachowicz/stachowicz.shtml, and Jenna Lang (http://jennomics.com/) at the University of California, Davis. The Seagrass Microbiome Project aims to integrate the long interest in seagrass ecology and ecosystem science with more recent work on microbiomes to produce a deeper, more mechanistic understanding of the ecology and evolution of seagrasses and the ecosystems on which they depend. Our studies of the community of microorganisms that live in and on seagrasses – the seagrass “microbiome” – will contribute to a broader understanding of host-microbe systems biology, and will benefit from ongoing University of Oregon research programs including the Microbial Ecology and Theory of Animals Center for Systems Biology (http://meta.uoregon.edu/) and the Biology and Built Environment Center (http://biobe.uoregon.edu/).
The position is available for 1 year with the possibility for renewal depending on performance. The start date is flexible. Please email questions regarding the position to Jessica Green (jlgreen).
To apply
A complete application will consist of the following materials:
(1) a brief cover letter explaining your background and career interests
(2) CV (including publications)
(3) names and contact information for three references
Submit materials to ie2jobs. Subject: Posting 14431
To ensure consideration, please submit applications by November 1, 2014, but the position will remain open until filled.
Women and minorities encouraged to apply. We invite applications from qualified candidates who share our commitment to diversity.
The University of Oregon is an equal opportunity, affirmative action institution committed to cultural diversity and compliance with the ADA. The University encourages all qualified individuals to apply, and does not discriminate on the basis of any protected status, including veteran and disability status.
Friday, November 21, 2014
Repeated, extremely biased ratio of M:F at meetings from SFB 680 "Evolutionary Innovations" group #YAMMM
Well, this is disappointing, to say the least - there is a conference coming up in July 2015 on "Forecasting Evolution": SFB 680 | Molecular Basis of Evolutionary Innovations at the Gulbenkian Foundation in Lisbon.
Here is the listed lineup of invited speakers:
- Andersson (Uppsala University), (NOTE I AM ASSUMING THIS IS DAN ANDERSSON)
- Trevor Bedford (Hutchinson Cancer Research Center),
- Jesse Bloom (Fred Hutchinson Cancer Research Center),
- Arup Chakraborty (MIT)
- Michael Desai (Harvard University),
- Michael Doebeli (University of British Columbia),
- Marco Gerlinger (Institute of Cancer Research, London,
- Michael Hochberg (CRNS, Montpellier),
- Christopher Illingworth (Cambridge University),
- Roy Kishoni (Harvard University),
- Richard Lenski (Michigan State University),
- Stanislas Leibler (Rockefeller University),
- Marta Luksza (IAS Princeton),
- Luke Mahler (University of California, Davis),
- Leonid Mirny (MIT),
- Richard Neher (MPI Tuebingen),
- Julian Parkhill (Sanger Institute),
- Colin Russell (University of Cambridge),
- Sohrab Shah (University of British Columbia),
- Boris Shraiman (UCSB),
- Olivier Tenaillon (Inserm Paris).
So I dug around a little bit. Here is another meeting from the same group at the University of Cologne - a group known as SFB 680. SFB 680: Molecular Ecology and Evolution: Cologne Spring Meeting 2012.
Sunday, November 16, 2014
Really cool: 3D printed microbes for the visually impaired
| Image from Nanowerk story. |
Tuesday, November 11, 2014
Job Announcement: Moore Foundation Program Associate
This seems like a potentially interesting job. I note - I love the Moore Foundation - and just about everything they are doing in science. Below is the email I recieved from the Project Lead Jon Kaye:
We have opened a search for a Program Associate. Details at the link below and attached. Please share with Bachelor’s and Master’s level individuals who may be interested. http://www.moore.org/about/careers?gnk=job&gni= 8a8725d0494f97e601495deb88ba30 cb We will also be opening a search for a PhD-level Program Officer position soon—stay tuned for details.
Full text of the announcement is below:
Sunday, November 02, 2014
Crosspost from #microBEnet: More scary than Halloween: this month in germophobia microbophobia
Crossposting this from the microBEnet blog where I originally posted it 10/31/14:
It seems that any time a holiday comes around in the US, the press starts to ramp up the writing of stories about evil microbes that are lurking all around us. And Halloween appears to be no exception. I am now planning on referring to this attitude as "microbophobia" rather than "germophobia" because to some "germ" implies pathogen and many of these stories fan the flames of fear about any kind of microbe not just pathogens. I note - the term microbophobia comes from some searches I did recently of Google books.
I was thinking of writing up yet another post trying to counter this excessive microbophobia but decided instead to just provide a collection of links to stories over the last month that have a distinctive microbophobia flavor. Mind you - there are real reasons to be afraid of some of the microbes circulating around these days. But the links below seem to me to be serious overkill.
It seems that any time a holiday comes around in the US, the press starts to ramp up the writing of stories about evil microbes that are lurking all around us. And Halloween appears to be no exception. I am now planning on referring to this attitude as "microbophobia" rather than "germophobia" because to some "germ" implies pathogen and many of these stories fan the flames of fear about any kind of microbe not just pathogens. I note - the term microbophobia comes from some searches I did recently of Google books.
I was thinking of writing up yet another post trying to counter this excessive microbophobia but decided instead to just provide a collection of links to stories over the last month that have a distinctive microbophobia flavor. Mind you - there are real reasons to be afraid of some of the microbes circulating around these days. But the links below seem to me to be serious overkill.
- KFOX 10/31/14: Special Assignment: How dirty is your cellphone?
- KENS 10/31/14: The most germ-laden thing in your home
- KFVS 10/30/14: Dirty candy: Trick-or-treating for candy or germs?
- KCCI Des Moines 10/30/14 Grinnell Regional Medical Center is using a robot to help fight infectious diseases like Ebola.
- Headlines and Global News 10/29/14: Germ-Free Homes: It's Possible, Even in the Wintertime
- Fox17 10/29/14 What part of your home is crawling with the most germs?
- Your Houston News 10/29/14: HAPPY HANDYMAN: Killing germs, bacteria much faster with Quick Clean Disinfectant
- Fox2Now 10/28/14: Blues bacterial infections, Halloween germs
- Cars.Com 10/28/14: With Flu Season Nearing Keep Germs at Bay in Car
- The Daily Telegraph: 10/28/14 Our filthy hospitals
- Bellingham Herald 10/27/14 Q&A: There are germs among us, and you can't be too safe
- Dothan Eagle 10/26/114 Scrubs will combat spread of germs
- Lifehacker 10/25/14: The Most Germ-Infested Item in the Office Might Be the Coffee Pot
- Shape Magazine 10/24/14: Confessions of A Germophobe
- Huffington Post: 10/17/14: This Terrifying Video Shows How Quickly Toilet Germs Spread To Your Mouth
- Daily Mail 10/14/14: How to Stay as Germ Free as Possible in the Toilets
- KLTV 10/10/14: Halloween Costume Shopping Brings Concern for Germs
- PSFK: Protective Transit Jacket for Germ-Free Commuting
- Wall Street Journal 10/1/14 Germs at the Office Are Often Found on Keyboards and at Coffee Stations
Friday, October 31, 2014
The flawed and offensive logic of "Academic Science Isn’t Sexist" in the @nytimes
OK. It is Halloween night and I am tired and need to get my kids to sleep. But someone on Twitter just pointed me to an opinion piece just out in the New York Times: Academic Science Isn’t Sexist - NYTimes.com and after reading it I felt I had to write a quick post.
The opinion piece is by Wendy M. Williams and Stephen J. Ceci and discusses work by them (and coauthors). In particular they discuss findings in a massive report "Women in Academic Science: A Changing Landscape" by Stephen J. Ceci, Donna K. Ginther, Shulamit Kahn, and Wendy M. Williams in Psychological Science in the Public Interest. I note - kudos to the authors for making this available freely and under what may be an open license and also apparently for making much of their data available behind their analyses.
The opinion piece and the associated article have a ton of things to discuss and ponder and analyze for anyone interested in the general issue of women in academic science. I am not in any position at this time to comment on any of the specific claims made by the authors on this topic. But certainly I have a ton of reading to do and am looking forward to it.
However, I do want to write about one thing - really just one single thing - that really bothers me about their New York Times article. I do not know if this was intentional on their part, but regardless I think there is a major flaw in their piece.
First, to set the stage -- their article starts off with the following sentences:
But the piece makes what to me appears to be a dangerous and unsupported connection. They lump together what one could call "career progression" topics (such as pay, promotion, publishing, citation, etc) with workplace topics (hostility and physical aggression against women). And yet, they only present or discuss data on the career progression issues. Yet once they claim to find that career progression for women in math heavy fields seems to be going well recently, they imply that the other workplace issues must not be a problem. This is seen in statements like "While no career is without setbacks and challenges" and "As we found, when the evidence of mistreatment goes beyond the anecdotal" and "leading many to claim that the inhospitable work environment is to blame."
Whether one agrees with any or all of their analyses (which again, I am not addressing here) I see no justification for their inclusion of any mention of hostile workplaces and physical agression against women. So - does this mean that a woman who does well in her career cannot experience physical aggression of any kind? Also - I note - I am unclear I guess in some of their terminology usage - is their use of the term "physical aggression" here meant to discount reports of sexual violence? This reminds me of the "Why I stayed" stories of domestic violence. Just because a women's career is doing OK does not mean that she did not experience workplace hostility or physical or sexual violence. I hope - I truly hope - that the authors did not intend to imply this. But whether they did or not, their logic appears to be both flawed and offensive.
UPDATE 1. November 1, 8:30 AM
Building a Storify about this.
UPDATE 2: Nov 3, 2014. Some other posts also criticizing the NY Times piece
The opinion piece is by Wendy M. Williams and Stephen J. Ceci and discusses work by them (and coauthors). In particular they discuss findings in a massive report "Women in Academic Science: A Changing Landscape" by Stephen J. Ceci, Donna K. Ginther, Shulamit Kahn, and Wendy M. Williams in Psychological Science in the Public Interest. I note - kudos to the authors for making this available freely and under what may be an open license and also apparently for making much of their data available behind their analyses.
The opinion piece and the associated article have a ton of things to discuss and ponder and analyze for anyone interested in the general issue of women in academic science. I am not in any position at this time to comment on any of the specific claims made by the authors on this topic. But certainly I have a ton of reading to do and am looking forward to it.
However, I do want to write about one thing - really just one single thing - that really bothers me about their New York Times article. I do not know if this was intentional on their part, but regardless I think there is a major flaw in their piece.
First, to set the stage -- their article starts off with the following sentences:
Academic science has a gender problem: specifically, the almost daily reports about hostile workplaces, low pay, delayed promotion and even physical aggression against women. Particularly in math-intensive fields like the physical sciences, computer science and engineering, women make up only 25 to 30 percent of junior faculty, and 7 to 15 percent of senior faculty, leading many to claim that the inhospitable work environment is to blame.This then sets the stage for the authors to discuss their analyses which leads them to conclude that in recent times, there are not biases against women in hiring, publishing, tenure, and other areas. Again, I am not in any position to examine or dispute their claims about these analyses - to either support them or refute them.
But the piece makes what to me appears to be a dangerous and unsupported connection. They lump together what one could call "career progression" topics (such as pay, promotion, publishing, citation, etc) with workplace topics (hostility and physical aggression against women). And yet, they only present or discuss data on the career progression issues. Yet once they claim to find that career progression for women in math heavy fields seems to be going well recently, they imply that the other workplace issues must not be a problem. This is seen in statements like "While no career is without setbacks and challenges" and "As we found, when the evidence of mistreatment goes beyond the anecdotal" and "leading many to claim that the inhospitable work environment is to blame."
Whether one agrees with any or all of their analyses (which again, I am not addressing here) I see no justification for their inclusion of any mention of hostile workplaces and physical agression against women. So - does this mean that a woman who does well in her career cannot experience physical aggression of any kind? Also - I note - I am unclear I guess in some of their terminology usage - is their use of the term "physical aggression" here meant to discount reports of sexual violence? This reminds me of the "Why I stayed" stories of domestic violence. Just because a women's career is doing OK does not mean that she did not experience workplace hostility or physical or sexual violence. I hope - I truly hope - that the authors did not intend to imply this. But whether they did or not, their logic appears to be both flawed and offensive.
UPDATE 1. November 1, 8:30 AM
Building a Storify about this.
UPDATE 2: Nov 3, 2014. Some other posts also criticizing the NY Times piece
- Emily Willingham: Academic science is sexist: We do have a problem here
- PZ Myers: Yay! Sexism in science is over!
- Red Ink: Let Me Fix That For You, New York Times
- Matthew Francis in Gailieo's Pendulum: No, academic science hasn’t overcome sexism (Nov 1, 2014)
UPDATE 3: Nov. 4, 2014. More posts about the NY Times piece
- Nick Desantis in the Chronicle for Higher Education: Op-Ed Called ‘Academic Science Isn’t Sexist’ Spurs Criticism (Nov 3, 2014)
- Athene Donald in the Guardian: Is the sexist scientific workplace really dead? (Nov 4, 2014)
Thursday, October 30, 2014
Rediscovering some critical terms of use in microbial discussions: #microbiomania and #microbophobia
Earlier this week I was trying to come up with a short term to use when referring to the "Overselling of the Microbiome" and related hype. And I came up with one I really really like: microbiomania. The term just captures the essence of hype about microbiomes to me I guess.
So - of course - the first thing to do was to see if anyone else used this term. And the number one thing I looked at was domain names. Nope. Microbiomania.Com and Microbiomania.Org are now mine. And then I started to search the interwebs. And surprsingly there was not much (in English at least). But some links showed up to books in Google Books with passages from > 100 years ago. And this is when the digging got to be fun. Here are some of the things I found.
1. A section from "The Medical Era"
When copying this section of the search results I discovered Google Books has an embed tool for Google Books though not sure how well it works: here is a try

Anyway - the text of this section of the book reads:
2. The Eclectic Medical Journal Volume 48
So - of course - the first thing to do was to see if anyone else used this term. And the number one thing I looked at was domain names. Nope. Microbiomania.Com and Microbiomania.Org are now mine. And then I started to search the interwebs. And surprsingly there was not much (in English at least). But some links showed up to books in Google Books with passages from > 100 years ago. And this is when the digging got to be fun. Here are some of the things I found.
1. A section from "The Medical Era"
When copying this section of the search results I discovered Google Books has an embed tool for Google Books though not sure how well it works: here is a try
Anyway - the text of this section of the book reads:
The Paris correspondent of the Chicago Tribune in a recent letter says We hear very little now of microbist or anti microbist theories Dr Koch's so called discovery is regarded with skepticism though not refuted The truth is his assertions are generally held to be not proven Dr Peters the favorite pupil of the great surgeon Dr Trousseau denounces what he calls microbiomania as a social danger and declares that the micro bians doctrine is vain sterile and objectionable in every way as both needlessly alarming and wrongly reassuringSo I guess there were some folks who did not like the Koch and his silly theories about germs.
2. The Eclectic Medical Journal Volume 48
Wednesday, October 29, 2014
CEO of Soylent goes even further off the deep end - going after his microbiome
Well, this is pretty deranged: Soylent CEO Is Lifehacking Water By Pissing In the Sink. Forget all the wackiness of Soylent and the idea of limiting water intake. And just look at the part of this on the micro biome
Hat tip to Andrea Kuszewski.
Feces are almost entirely deceased gut bacteria and water. I massacred my gut bacteria the day before by consuming a DIY Soylent version with no fiber and taking 500mg of Rifaximin, an antibiotic with poor bioavailability, meaning it stays in your gut and kills bacteria. Soylent's microbiome consultant advised that this is a terrible idea so I do not recommend it. However, it worked. Throughout the challenge I did not defecate.So - he took Rifaximin to kill his gut microbiome because he thought that would help him not defecate. And then because he did not defecate he concluded that the Rifaximin played some role in such anti-defecation? OMFG. This is both bad science and some, well, crazy a*s-sh*t. I - I - I - I just do not know what else to say.
Hat tip to Andrea Kuszewski.
Monday, October 27, 2014
Some suggestions for having diverse speakers at meetings
Been having a lot of discussions online in response to my post (Apparently, the National Academy of Sciences thinks only one sex is qualified to talk about alternatives to sex #YAMMM) tracking the awful gender ratio for speakers and session chairs at meetings run by the National Academy of Sciences in their Sackler series. Some people were asking what one can do to improve gender diversity at meetings so I thought I would post this which I was meaning to do anyway ...
-------------------------------------------------------
I wrote this in an email to a meeting organizer after I had turned down their invitation due to the imbalance in gender of the speakers (more about this another time --- this is not the same case as the one I wrote about here: Turning down an endowed lectureship because their gender ratio is too skewed towards males #WomenInSTEM).
-------------------------------------------------------
I wrote this in an email to a meeting organizer after I had turned down their invitation due to the imbalance in gender of the speakers (more about this another time --- this is not the same case as the one I wrote about here: Turning down an endowed lectureship because their gender ratio is too skewed towards males #WomenInSTEM).
Anyway, my colleague wrote a long and very helpful email to me after I withdrew from the meeting when I saw the speaker list. In the email she detailed things that her organization was trying to do to increase diversity of speakers at meetings. She ended it with this:
Thus, I take your comment to heart and wanted you to know that I care about this issues as well. I would love to hear how you balance these inequities at your meetings and learn as much as I can. Thank you for taking the time to bring this up I know how busy you are and appreciate your candor. Truly looking forward to more scientific exchanges and perhaps some education around gender issues.And I wrote back, quickly, without digging into the literature or all the posts in the world about this some quick suggestions which I think others might find useful. So here is my response - again - was not meant to cover all the things one can do - just examples:
Thanks so much for the response and I am really glad to see all you are trying to do in this area.
In terms of how we try to balance inequities at meetings I organize I would note a few simple things
Just some ideas off the top of my head.
- Do not try to invite only the famous people or the people doing the "top" work. This usually biases one towards more established researchers (as in, older) and this alas also usually is accompanied by distortion of diversity.
- DO try to invite people across the breadth of career stages. Meetings to me should not be only about getting the PIs whose labs are doing the best work to talk. It should also be about giving opportunities to junior researchers - PhD students, post docs and junior faculty who are doing exciting work - perhaps more focused or smaller scale - but nevertheless exciting. If one opens up a invited speaker list to people at diverse career stages one generally greatly increases the gender and ethnic diversity.
- DO try to invite people from diverse institutions - research universities, research institutes, companies, non profits, NGOs, the press, non research universities, and more.
- DO try to be flexible about times and dates for talks - I have found that women more than men have other commitments (e.g. kids) for which they cannot change dates of activities.
- DO try to provide child care assistance (as you are doing).
- DO try to make sure women are on the organizing committee See http://www.theatlantic.com/
technology/archive/2014/01/ the-easiest-possible-way-to- increase-female-speakers-at- conferences/282858/ - DO make sure to provide travel funds.
- DO try to include some talks on related areas that may not be the main theme of the conference. For example history of science and ELSI related topics increase the pool of women and speakers with diverse backgrounds which can be invited.
- DO ask the women who turn down invitations if they care to say why.
- DO commit to spending a decent amount of time searching for qualified female speakers. Sometimes there are people who fit ALL the goals of a meeting and they are just missed because women on average have lower public profiles than men doing the same type of work.
Jonathan
see also
http://www.stemwomen.net/jonathan-eisen/
and
http://phylogenomics.blogspot.com/p/posts-on-women-in- science.html
Friday, October 24, 2014
Apparently, the National Academy of Sciences thinks only one sex is qualified to talk about alternatives to sex #YAMMM
January 9-10, 2015
In the Light of Evolution IX. Clonal Reproduction: Alternatives to Sex
Organizers: Michel Tibayrenc, John C. Avise and Francisco J. Ayala
Beckman Center of the National Academies, Irvine, CA
Evolutionary studies of clonal organisms have advanced considerably in recent years, but are still fledgling. Although recent textbooks on evolution and genetics might give the impression that nonsexual reproduction is an anomaly in the living world, clonality is the rule rather than the exception in many viruses, bacteria, and parasites that undergo preponderant asexual evolution in nature. Clonality is thus of crucial importance in basic biology as well as in studies dealing with transmissible diseases.
This Colloquium will bring together specialists in various disciplines, including genetics, evolution, statistics, bioinformatics, and medicine. A balance will be sought between the various disciplines, including clonal animals and plants, animal and human cloning, pathogens, and cancer studies.
Registration is now open http://www.nasonline.org/programs/sackler-colloquia/upcoming-colloquia/ILE_IX_Clonal_Reproduction.html
Registration fee is $150.
Graduate students and postdoctoral researchers are eligible for discount fee of $100.
All meals, break and reception refreshments listed on the agenda are included in the registration fee.
For more information, contact sackler@nas.edu.
Could be interesting right? Alas, then, I clicked on the link. And I discovered the meeting could also be referred to as "Only one sex talks about alternatives to sex". Men are highlighted in yellow. Women highlighted in green. (Note - I am making some guesses as to gender but I think these are reasoably accurate).
Thursday, October 23, 2014
Microbiology Book for Kids: It's Catching by Jennifer Gardy and Josh Holinaty
A few days ago I wrote about how I wanted to share some information about what I have found to be good childrens' science books (based on reading books to my kids). Well, here is another one: It's Catching: The Infectious World of Germs and Microbes by Jennifer Gardy and Josh Holinaty.
by Jennifer Gardy and Josh Holinaty.
I first became aware of Jennifer Gardy's talents in making catchy microbe-themed kids material when she released the Youtube video "The A-Z of Epidemiology: germs from Anthrax to Zoonoses. A disturbing bedtime book for kids." which is simply awesome. (Note - great animation by Tom Scott):
I watched this video many many many times with my kids - always resulting in painful laughter and entertainment.
I should note that I am collaborating with Jennifer on at least one project (The Kitten Microbiome) and think she is a brilliant scientist and science communicator. But once I saw her "It's Catching" I realized she really could have a full career as a children's science book and video maker. It's Catching is both entertaining (like the video) but also educational with information on the history of microbiology and how microbes are studied. Definitely a good one if you are looking for fun and funny science and/or microbiology themed books for kids.
I first became aware of Jennifer Gardy's talents in making catchy microbe-themed kids material when she released the Youtube video "The A-Z of Epidemiology: germs from Anthrax to Zoonoses. A disturbing bedtime book for kids." which is simply awesome. (Note - great animation by Tom Scott):
I watched this video many many many times with my kids - always resulting in painful laughter and entertainment.
I should note that I am collaborating with Jennifer on at least one project (The Kitten Microbiome) and think she is a brilliant scientist and science communicator. But once I saw her "It's Catching" I realized she really could have a full career as a children's science book and video maker. It's Catching is both entertaining (like the video) but also educational with information on the history of microbiology and how microbes are studied. Definitely a good one if you are looking for fun and funny science and/or microbiology themed books for kids.
Wednesday, October 22, 2014
Microbe-themed art of the month: Seung-Hwan Oh portraits w/ mold
OK this is pretty cool (from a microbe-art-science point of view): An Artist Who Paints Portraits With Mold | WIRED. Seung-Hwan Oh "had to set up a micro-fungus farm in his studio" and he puts film in a warm wet environment (note to self - there could be a new human microbiome aspect of this project depending on what warm wet environment is chosen) and sometimes seeds the system with some mold. And then he lets nature do its work.
See more about his Impermanence works here. (Really - check out the works - they are wild).
At that site the work is described in the following way:
Hat tip to Kate Scow for posting about this on Facebook.
See more about his Impermanence works here. (Really - check out the works - they are wild).
At that site the work is described in the following way:
The visual result of the symbiosis between film matter and organic matter is the conceptual origin of this body of work. The process involves the cultivation of emulsion consuming microbes on a visual environment created through portraits and a physical environment composed of developed film immersed in water. As the microbes consume light-sensitive chemical over the course of months or years, the silver halides destabilize, obfuscating the legibility of foreground, background, and scale. This creates an aesthetic of entangled creation and destruction that inevitably is ephemeral, and results in complete disintegration of the film so that it can only be delicately digitized before it is consumed.Also see his Tumbl page where one can find many other images like this one:
Hat tip to Kate Scow for posting about this on Facebook.
Sunday, October 19, 2014
Kids' Microbiology Book Review: Germ Stories
I was going through some kids' books today and found quite a few that I thought were wonderful and thought - well - I should post about some of them. So that is what I am going to do.
The first I want to write about is Germ Stories by Arthur Kornberg with Illustrations by Adam Alaniz and Photos by Roberto Kolter.
by Arthur Kornberg with Illustrations by Adam Alaniz and Photos by Roberto Kolter.

I used to read it to my daughter all the time (she is two years older than my son) and then sometimes, when she was older, she would read it to my son. A few things I like about this book:
I added this book to a collection I am making via Amazon on "Microbiology Books for Kids". I will write about some of the other ones at another time.
UPDATE - Wanted other suggestions for good kids' microbiology themed books ...
The first I want to write about is Germ Stories

I used to read it to my daughter all the time (she is two years older than my son) and then sometimes, when she was older, she would read it to my son. A few things I like about this book:
- It is not all about pathogens - there are sections on yeast, penicillin, gut microbes and Myxococcus (although it is miswritten as Myxobacterium).
- Everything is done as poetry / songs. Some are cheesy, but my kids liked them.
- Each section on a different microbes has a little poem/song, a drawing, and a picture or two as well as a few mini facts (or I guess, micro facts).
- The material is a bit scary / gross at times but not too over the top.
Anyway - I definitely recommend it if you want a microbiology book that will be good for reading to and reading by kids.
UPDATE - Wanted other suggestions for good kids' microbiology themed books ...
Wednesday, October 15, 2014
Harvard, hope and hype: the sad reason behind overselling diabetes stem cell work - raising money
Earlier in the week I got all fired up - not in a good way - about a press release and news stories relating to a new paper from Doug Melton on a insulin producing STEM cell study
Exceptionally disappointed in Doug Melton of @HHMI for publishing new important diabetes paper as #closedaccess pic.twitter.com/DefoWwmiBR
— Jonathan Eisen (@phylogenomics) October 10, 2014
With a little more discussion I just got more angry
.@HHMINEWS and publishes the groundbreaking paper behind a paywall so online the elite can see it #closedaccess pic.twitter.com/ZPriyCdHvY
— Jonathan Eisen (@phylogenomics) October 10, 2014
.@David_Dobbs @Stephen_Curry @rpg7twit massive hype yet paywall is a big f#*$ you to those who care about diabetics pic.twitter.com/MErZi5pzL7
— Jonathan Eisen (@phylogenomics) October 11, 2014
@cryptogenomicon @phylogenomics @Stephen_Curry @David_Dobbs @rpg7twit the paper is available at
http://t.co/Rt2el9b9W5
— HHMI NEWS (@HHMINEWS) October 11, 2014
I was angry both about the overselling of the implications of the paper and the fact that the paper was not published in an open manner. This was despite the stated goals of HHMI which funds some of the Melton Lab work.
.@Stephen_Curry @David_Dobbs @rpg7twit only 1/5 pubs on Melton HHMI page http://t.co/L1p2BEKrQ6 is free #closedaccess pic.twitter.com/KQDY5ZLjuA
— Jonathan Eisen (@phylogenomics) October 11, 2014
Subscribe to:
Comments (Atom)
Most recent post
A ton to be thankful for -- here is one part of that - all the acknowledgement sections from my scholarly papers
So - it is another Thanksgiving Day and in addition to thinking about family, and football, and Alice's Restaurant, I also think a lot a...

-
Wow. Just wow. And not in a good way. Just got an email invitation to a meeting. The meeting is " THE FIRST ANNUAL WINTER Q-BIO ...
-
I have a hardback version of The Bird Way by Jennifer Ackerma n but had not gotten around to reading it alas. But now I am listening to th...
-
There is a spreading surge of PDF sharing going on in relation to a tribute to Aaron Swartz who died a few days ago. For more on Aaron ...