OK OK. Everyone, everywhere keeps telling me "Viruses - don't forget about the viruses" whenever I talk about microbial communities or discuss work we are doing in my lab. Yes, I say "I know, I know" but then I panic and think "We can't even deal with the diversity of bacteria and archaea out there. We generally ignore the microbial eukaryotes. How the hell are we going to include viruses too?" But, in the end, if we are going to study microBIOMEs (as in, ecosystems that include viruses), then, well, we need to include all the players.
(I note - as I am writing this, my son asked me what I am doing and I said I am writing about viruses. He got excited and sad "Write about Ebola").
What got me on my viral kick today? Well, I have been thinking about viruses a lot recently. And then I saw this paper: The ISME Journal - Diversity of viral photosystem-I psaA genes. And it reminded me of the largely unknown diversity of viruses out there in the world.
Anyway - I was going to write a longer post about viruses, but I have to go and do something with my son.
Saturday, December 27, 2014
Friday, December 26, 2014
Paper of possible interest: Exploring interactions of plant microbiomes
Just got alerted to this new (open access) paper that seems like it should be of interest to those working on plant microbiomes Scientia Agricola - Exploring interactions of plant microbiomes. It has some useful summaries of work that has been done on plant microbiomes.
Wednesday, December 24, 2014
Do preprints count for anything? Not according to Elife & G3 & some authors ..
Well, just got pointed to this paper: Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms | eLife by Martial Marbouty, Axel Cournac, Jean-François Flot, Hervé Marie-Nelly, Julien Mozziconacci, Romain Koszul. Seems potentially really interesting.
It is similar in concept and in many aspects to a paper we published in PeerJ earlier in the year (see Beitel et al., 2014 Beitel CW, Froenicke L, Lang JM, Korf IF, Michelmore RW, Eisen JA, Darling AE. (2014) Strain- and plasmid-level deconvolution of a synthetic metagenome by sequencing proximity ligation products. PeerJ 2:e415 http://dx.doi.org/10.7717/peerj.415.
Yet despite the similarities to our paper and to another paper that was formally published around the time of ours, this new paper does not mention these other pieces of work any where in the introduction as having any type of "prior work" relevance. Instead, they wait until late in their discussion:
Taking advantage of chromatin conformation capture data to address genomic questions is a dynamic field: while this paper was under review, two studies were released that also aimed at exploiting the physical contacts between DNA molecules to deconvolve genomes from controlled mixes of microorganisms (Beitel et al., 2014; Burton et al., 2014).Clearly, what they are trying to do here is to claim that since they paper was submitted before these other two (including ours) was published, that they should get some sort of "priority" for their work. Let's look at that in more detail. Their paper was received May 9, 2014. Our paper was published online May 27 and the other related paper by Burton et al. was published online May 22. In general, if a paper on what your paper is about comes out just after you submit your paper, while your paper is still in review, the common, normal thing to be asked to do is to rewrite your paper to deal with the fact that you were, in essence, scooped. But that does not really appear to be the case here. They are treating this in a way as "oh look, some new papers came out at the last minute and we have commented on them." The last minute would be in this case, 6 months before this new paper was accepted. Seems like a long time to treat this as "ooh - a new paper came up that we will add a few comments about".
But - one could quibble about the ethics and policies of dealing with papers that were published after one submitted one's own paper. From my experience, I have always had to do major rewrites to deal with such papers. But maybe E-Life has different policies. Who knows. But that is where things get really annoying here. This is because it was May 27 when our FINAL paper came out online at PeerJ. However, the preprint of the paper was published on February 27, more than two months before their paper was even submitted. So does this mean that the authors of this new paper do not believe that preprints exist? It is pretty clear on the web site for our paper that there is a preprint that was published earlier. Given what they were working on - something directly related to what our preprint/paper was about, one would assume they would have seen it with a few simple Google searches. Or a reviewer might have pointed them to it. Maybe not. I do not know. But either way, our preprint was published long before their paper was submitted and therefore I believe they should have discussed it in more detail.
Is this a sign that some people believe preprints are nothing more than rumors? I hope not. Preprints are a great way to share research prior to the delays that can happen in peer review. And in my opinion, preprints should count as prior research and be cited as such. I note - the Burton group in their paper in G3 also did not reference our preprint in what I consider to be a reasonable manner. They add some comments in their acknowledgements
While this manuscript was in preparation, a preprint describing a related method appeared in PeerJ PrePrints (Beitel et al. 2014a). Note added in proof: this preprint was subsequently published (Beitel et al. 2014b).Given that our preprint was published before their paper was submitted too, I believe that they also should have made more reference to it in their paper. But again, I can only guess that both the Burton and the Marbouty group just do not see preprints as being respectable scientific objects. That is a bad precedent to set and I think the wrong one too. And it is a shame. A preprint is a publication. No - it is not peer reviewed. But that does not mean it should not be considered part of the scientific literature in some way. I note - this new paper from the Marbouty group seems really interesting. Not sure I want to dig into it any deeper if they are going to play games with the timing of submission vs. published "papers" as part of how they are positioning themselves to be viewed as doing something novel.
Tuesday, December 23, 2014
Buzzfeed includes microbiomes in "77 facts" but alas, gets the facts (and the math) wrong
Well, I admit it - I clicked on this Buzzfeed link someone posted on Facebook - 77 Facts That Sound Like Huge Lies But Are Actually Completely True. There are some pretty funny and interesting things on the list. However, I note - I did not click on the link per se for entertainment. I clicked on it to see if there was anything about microbes on the list. And happily, there was. Alas, what there was, was, well, a bit wrong:
Let's start with #67. "There is 10 times more bacteria in your body than actual body cells". Well, alas, this is a nice bit of information. But it is not based on facts. See Peter Andrey Smiths wonderful article in the Boston Globe about this issue. The Buzzfeed article links to a 2010 Discover Magazine bit about this topic. So they were trying to use facts. But alas, that was not based on facts itself.
Then, let's go to #68. "And 90% of the cells that make us up of aren’t human but mostly fungi and bacteria.". So - I am at a bit of a loss on this. First, isn't this really just rehashing #67 with the addition of fungi to the story? Also - I note the math here is weird. #67 would lead one to conclude that 90.9% of the cells in your body are bacterial (10:1 = 10/11 = 90.9%). So then if one adds fungi to the picture the percent of cells in our bodies that are not human goes DOWN to 90%. How does that work exactly?
So not only do they get the 10x as many cells fact wrong, they do something really weird by basically repeating #67 and then doing some weird math.
Oh well, glad they have something on microbiomes. Too bad it came out a bit wrong.
Let's start with #67. "There is 10 times more bacteria in your body than actual body cells". Well, alas, this is a nice bit of information. But it is not based on facts. See Peter Andrey Smiths wonderful article in the Boston Globe about this issue. The Buzzfeed article links to a 2010 Discover Magazine bit about this topic. So they were trying to use facts. But alas, that was not based on facts itself.
Then, let's go to #68. "And 90% of the cells that make us up of aren’t human but mostly fungi and bacteria.". So - I am at a bit of a loss on this. First, isn't this really just rehashing #67 with the addition of fungi to the story? Also - I note the math here is weird. #67 would lead one to conclude that 90.9% of the cells in your body are bacterial (10:1 = 10/11 = 90.9%). So then if one adds fungi to the picture the percent of cells in our bodies that are not human goes DOWN to 90%. How does that work exactly?
So not only do they get the 10x as many cells fact wrong, they do something really weird by basically repeating #67 and then doing some weird math.
Oh well, glad they have something on microbiomes. Too bad it came out a bit wrong.
Sunday, December 21, 2014
Crosspost - papers for sale
Crossposting from ICIS Blog (even though this is short - always good to reveal where things are posted first).
Well, this is much more elaborate than I could ever have imagined: For Sale: “Your Name Here” in a Prestigious Science Journal – Scientific American. Seems that there are services out there to help people write, in essence, bogus scientific papers filled with pithy somewhat reasonable sounding phrases about certain topics. Seems we could all use some more comprehensive full text analyses of papers to try and flag such activities.
Well, this is much more elaborate than I could ever have imagined: For Sale: “Your Name Here” in a Prestigious Science Journal – Scientific American. Seems that there are services out there to help people write, in essence, bogus scientific papers filled with pithy somewhat reasonable sounding phrases about certain topics. Seems we could all use some more comprehensive full text analyses of papers to try and flag such activities.
Thursday, December 18, 2014
Great idea from Nicole King - Lists of Women Speakers - more examples wanted
Just been pointed to this compilation from Nicole King (a brilliant professor at Berkeley): Lists of Women Speakers | The King Lab.
She has put together a collection of a few links:
She has put together a collection of a few links:
As a resource for those who are interested in increasing the number of female speakers at their scientific conferences, seminar series, etc., I offer here a compendium of female speakers from diverse fields. I've started with lists that I could find easily, but if you know of additional lists, please feel free to send along the links. I would also be happy to maintain similar lists for underrepresented minorities if any such lists exist.These are the lists she maintained:
General
- University of Minnesota, Distinguished Scientists and Engineers from Underrepresented Groups
- Women scientists that give awesome seminars
- Celebrating Women in Science Symposium
- Indiana University Joan Wood Lecture Series
Field-specific lists
- American Society of Cell Biology WICB Speaker Referral Service
- Neuroscience: Annie's List
- Theoretical/computational chemistry, material science, and biochemistry
- Synthetic Biology
- Prominent ecologists
- Geoscientists
- American Physical Society Women Speakers List
Are people aware of any other relevant lists like this? Please share I will compile ...
Wednesday, December 10, 2014
New lab paper: The microbes we eat: abundance and taxonomy of microbes consumed in a day’s worth of meals for three diet types
A new paper out from my lab (with Jenna Lang as the 1st author and in collaboration with Angela Zivcovic from the UC Davis Food For Health Initiative and the Department of Nutrition): The microbes we eat: abundance and taxonomy of microbes consumed in a day’s worth of meals for three diet types. The work in the paper focuses on characterizing the abundance and taxonomy of microbes in food from three model diets.
Basically, Angela prepared meals for these three diets
Lots of interesting things reported in the paper (read it, I insist). I note - this is a demonstration project in a way - trying to get the community and others to think about the source pools of microbes that come into our system from our food. It is by no means comprehensive or conclusive (read the caveats section of the paper). Congrats to Jenna and Angela for all their hard work. Anyway - the paper is Open Access in PeerJ. Eat it up.
UPDATE: Some press and blog coverage
Basically, Angela prepared meals for these three diets
Food was purchased and prepared in a standard American home kitchen by the same individual using typical kitchen cleaning practices including hand washing with non-antibacterial soap between food preparation steps, washing of dishes and cooking instruments with non-antibacterial dish washing detergent, and kitchen clean-up with a combination of anti-bacterial and non-antibacterial cleaning products. Anti-bacterial products had specific anti-bacterial molecules added to them whereas “non-antibacterial” products were simple surfactant-based formulations. The goal was to simulate a typical home kitchen rather than to artificially introduce sterile practices that would be atypical of how the average American prepares their meals at home. All meals were prepared according to specific recipes (from raw ingredient preparation such as washing and chopping, to cooking and mixing).And then she blended them and we characterzied the microbial communities in the blended samples:
After food preparation, meals were plated on a clean plate, weighed on a digital scale (model 157W; Escali, Minneapolis, MN), and then transferred to a blender (model 5,200; Vita-Mix Corporation, Cleveland, OH) and processed until completely blended (approximately 1–3 min). Prepared, ready to eat foods that were purchased outside the home were simply weighed in their original packaging and then transferred to the blender. 4 mL aliquots of the blended meal composite were extracted from the blender, transported on dry ice and then stored at −80 °C until analysis. The following analyses were completed using these meal composite samples: (1) total aerobic bacterial plate counts, (2) total anaerobic bacterial plate counts, (3) yeast plate counts, (4) fungal plate counts, and (5) 16S rDNA analysis for microbial ecology.And Jenna Lang coordainted the sequence analysis and then Angela and Jenna (with some help here and there from me) coordianted the analysis of the different microbial data and the writing of the paper.
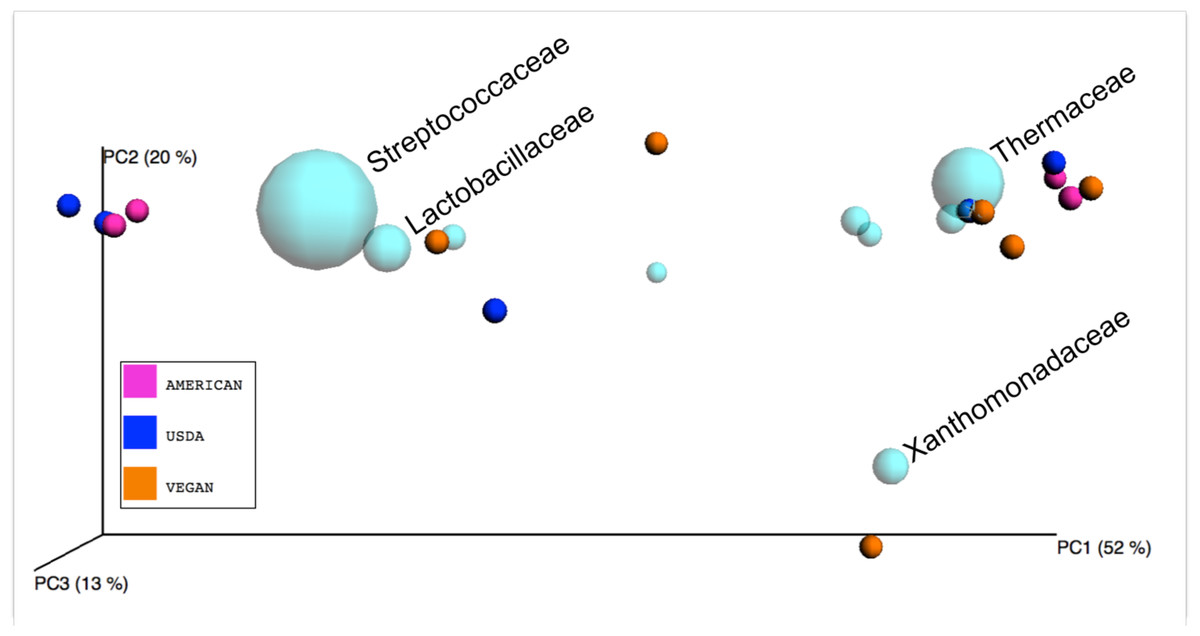 | ||
| Figure 5: Biplot of taxa in sample PCoA space. |
Lots of interesting things reported in the paper (read it, I insist). I note - this is a demonstration project in a way - trying to get the community and others to think about the source pools of microbes that come into our system from our food. It is by no means comprehensive or conclusive (read the caveats section of the paper). Congrats to Jenna and Angela for all their hard work. Anyway - the paper is Open Access in PeerJ. Eat it up.
UPDATE: Some press and blog coverage
- Kevin Bonham on SciAM Blogs: The Microbes in Your Kitchen (Or in your Starbucks mocha)
- Maddie Stone on Motherboard: Here’s How Many Microbes You Eat in a Single Day
- “This paper will hopefully convince the powers that be that the next study, actually looking at how different foods or diets with differing microbes affect people, should be funded soon,” Zivkovic told me.
- Nathan Gray: The microbes we eat: Study investigates bacteria in different diets
- Ross Pomeroy on Real Clear Science: This Is How Many Microbes You'll Eat Today
- Improbable Research: What’s Eating You, and/or Vice Versa: Microbes
- Marissa Fessenden at Smithsonian: You Eat Millions...Even Billions!...of Microbes Every Day
Saturday, December 06, 2014
Some history of hype regarding the human genome project and genomics
Just taking some notes here - relates to a discussion going on online. Would love pointers to other references relating to hype and the human genome project (including references that think it was not overhyped). I note - see some of my previous posts about this issue including: Human genome project oversold? sure but lets not undersell basic science and various Overselling Genomics awards.
Here are some things I have found:
White House press conference on announcing completetion of the human genome
The Human Genome Project: Hype meets reality
NOVA: Nature vs. Nurture Revisited
The human genome project, 10 years in: Did they oversell the revolution? in the Globa and Mail by Paul Taylor referring to: "Deflating the Genomic Bubble"
Also see Genomic Medicine: Too Great Expectations? by PP O Rourke
Also Has the Genomic Revolution Failed?
And Human genome 10th anniversary. Waiting for the revolution.
Science communication in transition: genomics hype, public engagement, education and commercialization pressures.
The Medical Revolution in Slate.
A Decade Later, Genetic Map Yields Few New Cures in the New York Times.
NNB report: Ten years later, Harvard assesses the genome map where regarding Eric Lander:
From Great 15-Year Project To Decipher Genes Stirs Opposition in the Times June 1990
From SCIENTIST AT WORK: Francis S. Collins; Unlocking the Secrets of the Genome
From READING THE BOOK OF LIFE: THE DOCTOR'S WORLD; Genomic Chief Has High Hopes, and Great Fears, for Genetic Testing June 2000 in the NY Times
The story goes through some predictions Francis Collins made for the future in a talk. These included:
Here are some things I have found:
White House press conference on announcing completetion of the human genome
Genome science will have a real impact on all our lives -- and even more, on the lives of our children. It will revolutionize the diagnosis, prevention and treatment of most, if not all, human diseases. In coming years, doctors increasingly will be able to cure diseases like Alzheimer's, Parkinson's, diabetes and cancer by attacking their genetic roots. In fact, it is now conceivable that our children's children will know the term cancer only as a constellation of stars.Collins et al. New Goals for the U.S. Human Genome Project: 1998–2003
The Human Genome Project (HGP) is fulfilling its promise as the single most important project in biology and the biomedical sciences— one that will permanently change biology and medicine.Human Genome -The Biggest Sellout in Human History
The Human Genome Project: Hype meets reality
NOVA: Nature vs. Nurture Revisited
After a decade of hype surrounding the Human Genome Project, punctuated at regular intervals by gaudy headlines proclaiming the discovery of genes for killer diseases and complex traits, this unexpected result led some journalists to a stunning conclusion. The seesaw struggle between our genes (nature) and the environment (nurture) had swung sharply in favor of nurture.
The human genome project, 10 years in: Did they oversell the revolution? in the Globa and Mail by Paul Taylor referring to: "Deflating the Genomic Bubble"
Also see Genomic Medicine: Too Great Expectations? by PP O Rourke
Also Has the Genomic Revolution Failed?
And Human genome 10th anniversary. Waiting for the revolution.
Science communication in transition: genomics hype, public engagement, education and commercialization pressures.
The Medical Revolution in Slate.
A Decade Later, Genetic Map Yields Few New Cures in the New York Times.
In announcing on June 26, 2000, that the first draft of the human genome had been achieved, Mr. Clinton said it would “revolutionize the diagnosis, prevention and treatment of most, if not all, human diseases.”
At a news conference, Francis Collins, then the director of the genome agency at the National Institutes of Health, said that genetic diagnosis of diseases would be accomplished in 10 years and that treatments would start to roll out perhaps five years after that.
Bursting the genomics bubble - Nature 2010
Has decoding the genome lived up to hype? BBC - 2010
The ethics of expectations: biobanks and the promise of personalised medicine.
The ethics of expectations: biobanks and the promise of personalised medicine.
NNB report: Ten years later, Harvard assesses the genome map where regarding Eric Lander:
At the same time, , he said genomic research has “gone so much faster than I would have imagined.” He cited " an explosion of work that will culminate, I think in the next five years, in a pretty comprehensive list of all the target that lead to different kinds of cancers and give us a kind of roadmap for finding the Achilles heel of cancers for therapeutics and diagnostics."while at the same time he blamed the press for the hype
From Great 15-Year Project To Decipher Genes Stirs Opposition in the Times June 1990
'Our project is something that we can do now, and it's something that we should do now,'' said Dr. James D. Watson, a Nobel laureate who heads the National Center for Human Genome Research at the National Institutes of Health. ''It's essentially immoral not to get it done as fast as possible.''
- Note the article has many complaining about the hype in the genome project even then ..
From SCIENTIST AT WORK: Francis S. Collins; Unlocking the Secrets of the Genome
And, Dr. Collins adds, there is nothing more important in science and medicine than the project he heads
Dr. Collins predicts that within 10 years everyone will have the opportunity to find out his or her own genetic risks, to know if cancer or heart attacks or diabetes or Alzheimer's disease, for example, lies in the future.
From READING THE BOOK OF LIFE: THE DOCTOR'S WORLD; Genomic Chief Has High Hopes, and Great Fears, for Genetic Testing June 2000 in the NY Times
The story goes through some predictions Francis Collins made for the future in a talk. These included:
- BY 2010, the genome will help identify people at highest risk of particular diseases, so monitoring efforts can focus on them.
- In cancer, genetic tests will identify those at highest risk for lung cancer from smoking. Genetic tests for colon cancer will narrow colonoscopy screening to people who need it most. A genetic test for prostate cancer could lead to more precise use of the prostate specific antigen, or P.S.A., test by identifying those men in whom the cancer is most likely to progress fastest. Additional genetic tests would guide treatment of breast and ovarian cancer.
- Three or four genetic tests will help predict an individual's risk for developing coronary artery disease, thus helping to determine when to start drugs and other measures to reduce need for bypass operations.
- Tests predicting a high risk for diabetes should help encourage susceptible individuals to exercise and control their weight. Those at higher risk might start taking drugs before they develop symptoms.
- BY 2020, doctors will rely on individual genetic variations in prescribing new and old drugs and choosing the dose. Pharmaceutical companies will take a second look at some drugs that were never marketed, or were taken off the market, because some people who took them suffered adverse reactions. It will take many years to develop such drugs and tests.
- Cancer doctors will use drugs that precisely target a tumor's molecular fingerprint. One such gene-based designer drug, Herceptin, is already marketed for treating advanced breast cancer.
- The genome project holds promise for the mental health field. ''One of the greatest benefits of genomic medicine will be to unravel some biological contributions to major mental illnesses like schizophrenia and manic depressive disease'' and produce new therapies, Dr. Collins said.
Friday, December 05, 2014
Some notes on "Citations for Sale" about King Abdulaziz University offerring me $$ to become an adjunct faculty
There is a news story in the Daily Cal titled "Citations for Sale" by Megan Messerly about King Abdullah University King Abdulaziz University trying to pay researchers who are highly cited to become adjunct professors there to boost their rankings. This article stemmed from a blog post by Lior Pachter. I was interviewed by the Daily Cal reporter about this because I had sent Lior some of the communications I had had with people from KAU where they tried to get me to do this.
I am posting here some of the email discussions / threads that I shared with Lior and Megan.
Thread #1.
Here is one thread of emails in which KAU tried to get me to become an Adjunct Professor. I have pulled out the text of the emails and removed the senders ID just in case this would get him in trouble.
Received this email 3/6/14
Dear Prof Jonathan,
How are you? I hope every thing is going well. I hope to share work with you and initiate a collaboration between you and me in biology department, Kind Abdulaziz university . My research focuses on (redacted detail). I hope that you would agree .
Redacted Name,Ph.DMy response:
Assistant Professor, Faculty of Sciences, King Abdulaziz University, Jeddah, KSA.
What kind of collaboration are you imagining?His response:
Hi Prof Jonathan,
Let me to explain that the king abdulaziz university initiated a project which is called Highly Cited Professor ( HiCi). This project can provide a contract between you and our university and from this contract you can get 7000 US$ as a monthly salary .So this project will allow you to generate two research proposal between you and a research stuff here in order to make an excellent publications in high quality journals as you always do.
I hope that I was clear. I' m looking forward to hear from you. Finally, I think that a very good chance to learn from you.Another email from him:
Dear prof Jonathan,
I' d like to tell that Prof Inder Verma will come tomorrow to our university as a highly cited professor and he also signed a contract with us. At March 28 Prof Paul Hebert will come to our university and we actually generated two projects with Prof Paul. I hope you trust me and you call Prof Inder and Paul to be sure.From me:
I trust you - just am very busy so not sure if this is right for meFrom him:
Sent from my iPhone
You will come to our university for just two visits annually and each visit will take only one week. Take you time to think. ByeAnother email from him
Seat Dr Jonathan,
What is your decision?
Subscribe to:
Comments (Atom)
Most recent post
A ton to be thankful for -- here is one part of that - all the acknowledgement sections from my scholarly papers
So - it is another Thanksgiving Day and in addition to thinking about family, and football, and Alice's Restaurant, I also think a lot a...

-
Wow. Just wow. And not in a good way. Just got an email invitation to a meeting. The meeting is " THE FIRST ANNUAL WINTER Q-BIO ...
-
I have a hardback version of The Bird Way by Jennifer Ackerma n but had not gotten around to reading it alas. But now I am listening to th...
-
There is a spreading surge of PDF sharing going on in relation to a tribute to Aaron Swartz who died a few days ago. For more on Aaron ...