The Tree of Life
Blog of Jonathan A. Eisen, Professor at U.C. Davis.
Tuesday, January 30, 2024
My Ode to Yolo Bypass
Sunday, December 31, 2023
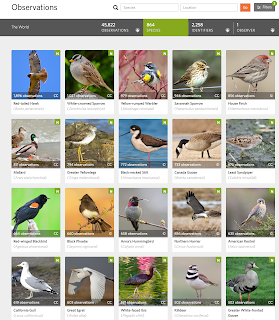
Four years - 1461 straight days - of iNaturalist observations
Well, today is a big day I guess. I just posted an observation to iNaturalist of a hummingbird in my backyard.
https://www.inaturalist.org/observations/195397005
Not that big a deal unless you are living in some place where it is really cold and you are jealous we have hummingbirds in our yard in Davis, California on December 31.
But the context here is important.
This entry completes a somewhat crazy run. I have made iNaturalist observations every day for the last four years. That is, every day for the last 1461 days.
It all started in January 2020. Before the pandemic reared its ugly head. I decided it might be fun to try and be more regular about iNaturalist postings. I had gotten into posting my observations there during 2018 and 2019 and then I saw on Twitter some people referring to an iNaturalist "Streak Finder" where you could look up how many days in a row someone posted observations.

And many people were discussing how many they had. And my max # at the time was OK (max of 16) but not particularly high.
And then I kind of forgot about it. But I was inspired by conversations with Laci Gerhart and I decided I would try to do an entry every day for all of 2020.
https://twitter.com/phylogenomics/status/1214614081329487872
And things of course got complicated quickly. First major complication -- I got called for Jury Duty and was selected as an alternate for a pretty serious case that in the end ended up lasting 3-4 weeks.
And I was pretty proud when I got through month 1 - January 2020. I posted a thread on Twitter about this.


I also posted this on my blog here.
One of the reasons I started doing this attempt to post every day was that I was hoping to go on a lot of work trips and to post every day on those trips and this would give me something to do while on the road.
In February I went to the AAAS Meeting in Seattle and on breaks wandered around and took pics and made lots of postings. Plus I got to hang out with an old friend, Carl Bergstrom, and we wandered around one day looking for birds
And things seemed good. Except for one thing. While at AAAS, I decided to avoid indoor crowded places because of the growing reports that this new respiratory disease had shown up in the US. And, well, this would be my last trip involving planes until a few weeks ago. So much for my plan to use iNaturalist as a way to help me not sit in hotels / conference centers while on trips.And there were many many struggles in life generally for me and many others as the pandemic spread. But somehow through this I managed to keep making observations. In fact, I would say without a doubt the mission to make observations helps keep me semi sane during some dark times.
And then I found myself on 12/31/2020. I had done it. One whole year of daily observations.
https://twitter.com/phylogenomics/status/1344873226464161792
And of course lots more happened over the next year. But I made it to 12/31/2021 and had made observations for another whole year.

And fast forward to today. I have made it to four years of daily observations. 1461 days. Through health issues. And the ongoing pandemic. And all sorts of complications. One thing that helped me in life was my mission to make some sort of nature observation I could post every day.
Thursday, September 28, 2023
Notes on My Meningioma and Gamma Knife Treatment
So - on 9/27 I had Gamma Knife treatment at UC Davis for a posterior fossa meningioma that was first found by MRI about 6 years ago. I am going to be collecting my various posts here on the saga. I am also going to post separate posts here about the saga but just wanted to collect some of the other posts for now.
Thursday, September 21, 2023
Sanger DNA sequencing services
Sanger DNA Sequencing Services
So I posted a request to multiple social media sites:
Question - are there good / cheap services out there to do Sanger sequencing (e.g., for 16S / ITS for taxonomic identification of cultures)? The local Sanger operation we were using shut down.
I posted to the following sites:
These are the places that were recommended the most
These had single suggestions
- Macrogene
- MR DNA
- Integrated Genomics
- Functional Bio
- U Az.
- CosmosID
- Laragen
- Retrogen
- IMR
- Functional Biosciences
These are how the suggestions mapped to the different sites:
- Eurofins
- McLab in south SF
- QB3
- Macrogene
- MR DNA
- Laragen
- Retrogen
- IMR
- Functional Biosciences
- Eurofins
- Quintara
- Functional Bio
- Gene Wiz (now Azenta)
- UC Berkeley
- U Az.
- Blue Sky
- Mastodon
- McLab
- Quintara
Thursday, August 24, 2023
And today in one of the stranger uses of my work ... "130 Academic Words Ref from "Jonathan Eisen: Meet your microbes | TED Talk""
Well, this is certainly a bit wacky.
I was, well, Googling myself today and found this.
130 Academic Words Ref from "Jonathan Eisen: Meet your microbes | TED Talk"
Most recent post
My Ode to Yolo Bypass
Gave my 1st ever talk about Yolo Bypass and my 1st ever talk about Nature Photography. Here it is ...

-
See Isolation and sequence-based characterization of a koala symbiont: Lonepinella koalarum Paper based on PhD thesis work of Katie Dahlha...
-
Just got this press release by email. I am sick of receiving dozens of unsolicited press releases, especially those in topics not related ...
-
I have a new friend in Google Scholar Updates I have written about the Updates system before and if you want more information please see...